GGGenome Package Version

High-throughput Genome Search Database
– Now Available Offline!
Nucleic acid therapeutics are short, typically around 20 bases, making it difficult to perform accurate searches using conventional base-search tools like BLAST.
Furthermore, depending on the target site of the nucleic acid therapeutic, the base sequence of pre-mRNA is required in addition to mRNA sequences, yet no well-maintained database existed for this purpose.
GGGenome is an accurate and comprehensive search system based on algorithms specialized for short nucleotide sequences. It includes the D3G database, which contains organized pre-mRNA nucleotide sequences. It has a proven track record in safety assessments for previously approved nucleic acid therapeutics.
The GGGenome Package Version is a system that adds features required for drug discovery and development to the functionality of the online version.
Furthermore, depending on the target site of the nucleic acid therapeutic, the base sequence of pre-mRNA is required in addition to mRNA sequences, yet no well-maintained database existed for this purpose.
GGGenome is an accurate and comprehensive search system based on algorithms specialized for short nucleotide sequences. It includes the D3G database, which contains organized pre-mRNA nucleotide sequences. It has a proven track record in safety assessments for previously approved nucleic acid therapeutics.
The GGGenome Package Version is a system that adds features required for drug discovery and development to the functionality of the online version.
Feature
 |
 |
 |
| Ultrafast Search of Short-base Sequences | Accurate Mismatch, Indel, and Ambiguity Search | Trust and Proven Track Record in Safety Assessment of Nucleic Acid Therapeutics |
| Nucleic acid therapeutics often have short sequences of ~20 bases. GGGenome enables efficient safety evaluation. | Perform essential searches for nucleic acid drug safety evaluation with precision and ease. | For nucleic acid therapeutic approval applications, this serves as a reliable source of search results. |
 |
 |
 |
 |
| Offline Search Capability | Eased Search Restrictions | Highlighting Featured Gene | Batch Search Made Easy with Search Scripts |
| Operates via USB (SSD), preventing confidential information leak. | Online search has restrictions to reduce server load, but Offline version has been eased, enabling efficient searching. | Easily extract only the important results from vast search results. | Complexed operations such as multiple searches can be assigned at once from the UI screen, enabling anyone to search efficiently. |
Features updated in the 2025 Edition
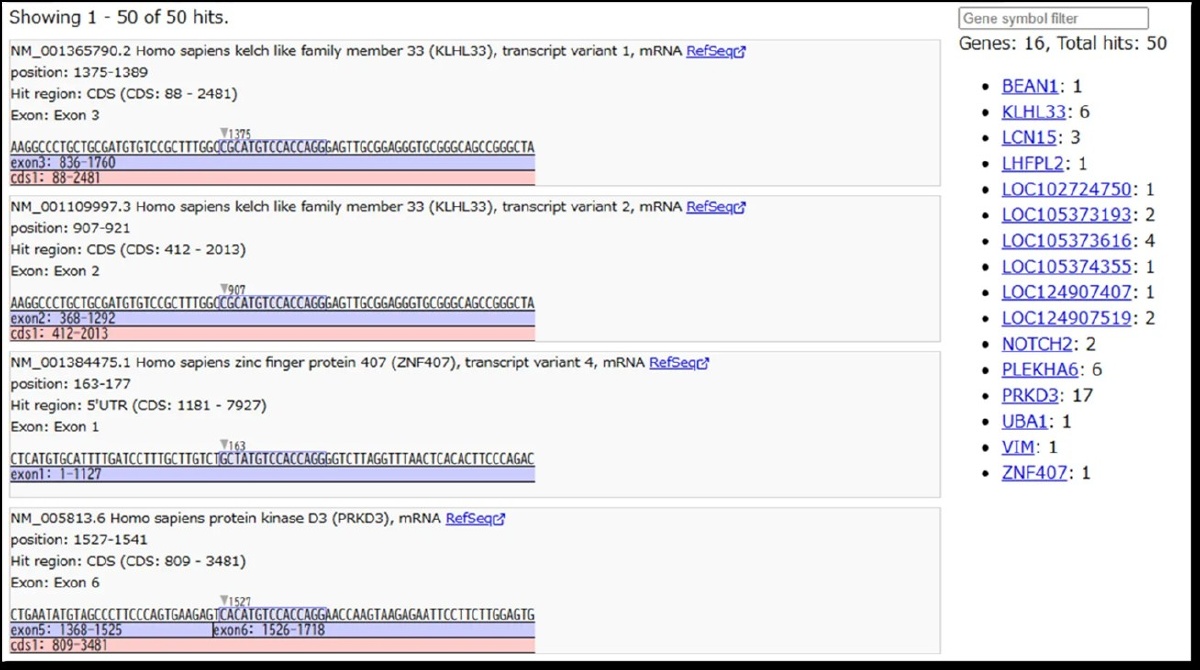
Original Search UI Screen has been added. Previously, off-target candidates were identified based on nucleotide sequences similar to those of nucleic acid drug targets.
With this version, users can now narrow down results by candidate gene name and understand where similar sequences occur within genes visually and intuitively (in coding regions, exons, or junctions). Search results can be output by selecting the required information.
With this version, users can now narrow down results by candidate gene name and understand where similar sequences occur within genes visually and intuitively (in coding regions, exons, or junctions). Search results can be output by selecting the required information.
List of bundled databases
GGGenome Drug Discovery Basic Pack 2025 Edition
Search Data (9 Species, 41 Databases)
D3G / RefSeq are all the latest versions
Search Data (9 Species, 41 Databases)
D3G / RefSeq are all the latest versions
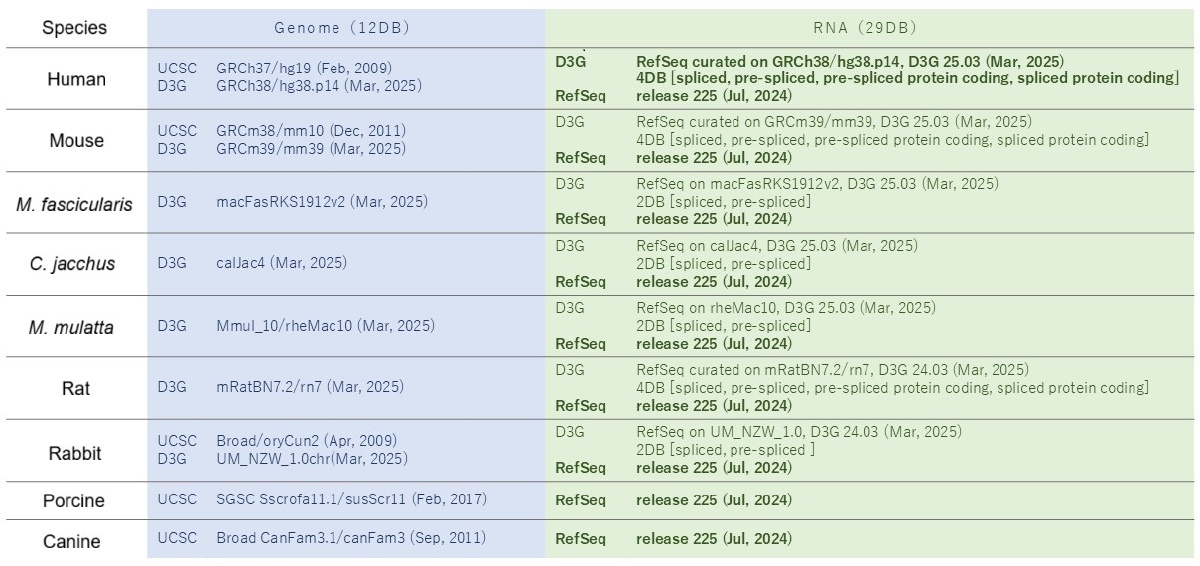
D3G was developed under the Kawai Group in Program for Promoting Platform of Genomics based Drug Discovery, AMED, Japan (FY2017–2022). It is currently maintained by the Kawaji Group's “Research on Establishing Infrastructure for Exploring Novel Drug Targets Using Genome Information” (FY2023-). It is widely used in nucleic acid-based drug discovery and regulatory affairs, accessible via the GGGenome web version.
Options
• Optional Databases: You can add databases available for web-based GGGenome published by DBCLS.
• Custom Databases: You can add your own nucleotide sequence databases.
• Custom Databases: You can add your own nucleotide sequence databases.
System Requirements
This product is compatible with 64-bit versions of Windows.
✓ Windows 11 Pro version 21H2 or later, or Enterprise/Education version 21H2 or later
✓ Windows 10 Pro 21H1 or later, or Enterprise/Education 20H2 or later
✓ Hyper-V must be enabled (may be mutually exclusive with virtualization software like VMware)
✓ Memory: 8GB or more (At least 4GB must be allocated to Docker for Windows)
✓ Windows 11 Pro version 21H2 or later, or Enterprise/Education version 21H2 or later
✓ Windows 10 Pro 21H1 or later, or Enterprise/Education 20H2 or later
✓ Hyper-V must be enabled (may be mutually exclusive with virtualization software like VMware)
✓ Memory: 8GB or more (At least 4GB must be allocated to Docker for Windows)









